Translation
Translation in another of processes of transferring genetic information in cell, following after transcription. During translation, a cell “reads” the information in a messenger RNA (mRNA) and uses it to build a protein.
DNA is long double stranded molecule that is made out of 4 nucleotides - A, T, C, G. Messenger RNA (mRNA) is shorter single stranded molecule also made out 4 nucleotides, but this time it is A, U, C, G. This U (uracil) nucleotide is RNA equivalent to T (thymine) nucleotide in DNA. Proteins are also single stranded molecules but they are not made out of nucleotides. This is why process of producing protein based on RNA is called translation - we are translating into a completely different kind of chemical. Building blocks of proteins are called amino acids. There are 20 different amino acids that are used in living organisms that we know. There is many more amino acids as chemical molecules, but they’re 20 that are used in in life.
Genetic code
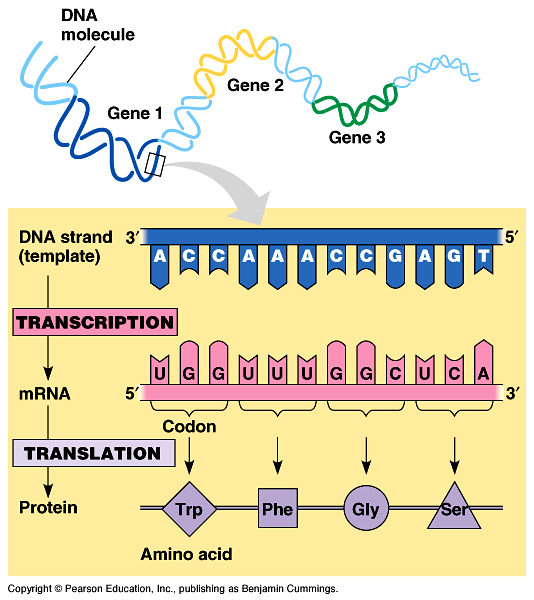
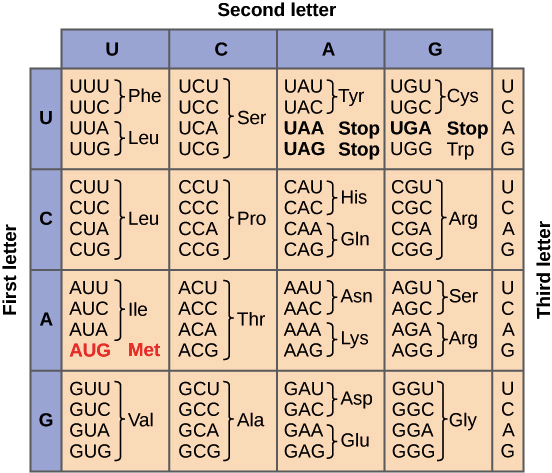
mRNA has a signal that needs to be translated into protein. This mRNA is made out of 4 letters - A, U, C, G. We want to translate this into a protein that can be build out out 20 different amino acids. We need some mapping between 4 nucleotides and 20 amino acids. Obviously, we cannot do one-to-one mapping as we did with DNA to RNA transcription. The way how life got around this is to use triplets (kmers where k=3). Combining three RNA nucleotides together, we can make up to 64 different combinations (three position, 4 possible nucleotides, 4*4*4 = 64). These 64 possible combinations are used to encode 20 different amino acids plus some control signals (such as stop signal). This dictionary of triplets (called codons in here) versus amino acids is called the genetic code.
Genetic code is often shown in table like the one below. On the left there is the 1st letter, then on the top is the 2nd letter and then each one of the cells has four triplets corresponding to each possibility at 3rd letter.
Degeneration
We have 20 (or 21 if we count Stop) amino acids that are encoded by 64 codons. This means that more that one codon can encode the same amino acid. This characteristic of genetic code is called degeneration and it allows to have a little bit of variability in RNA sequence and still get the same amino acid. However, this variability is not random - 3rd letter is most likely to be degenerated. We can see this in table of genetic code - blocks that have the same first two letters tend to encode the same amino acid regardless of the 3rd letter.
The fact that this code has three positions and the first two positions seem to be much more important than the third one, creates an effect in which if we look at the evolutionary conservation of protein-coding sequences, we would see that the 1st and 2nd letter tend to be very conservative, and then the third letter tends to be much less conserved. Change in the 3rd letter will probably not affect the protein that is being produced.
Universality
The genetic code is universal. With a few exceptions, virtually all species use the same genetic code for protein synthesis. Conservation of codons means that mRNA encoding the globin protein in horses could be transferred to a tulip cell, and the tulip would synthesize horse globin. That there is only one genetic code is powerful evidence that all of life on Earth shares a common origin, especially considering that there are about 1084 possible combinations of mapping between 20 amino acids and 64 triplet codons.
Special codons
On top of encoding for amino acids, there are also some regulatory signals encoded in mRNA. Codon AUG encodes amino acid Methionine, but it also has its second role as the start codon. All RNA sequences encoding for proteins start with AUG. There can be other AUGs within the sequence that encode for amino acid Methionine. Second very important regulatory signal is stop codon. There are three stop codons - UAA, UAG and UGA - that encode the end of the protein. No amino acid is associated with them.
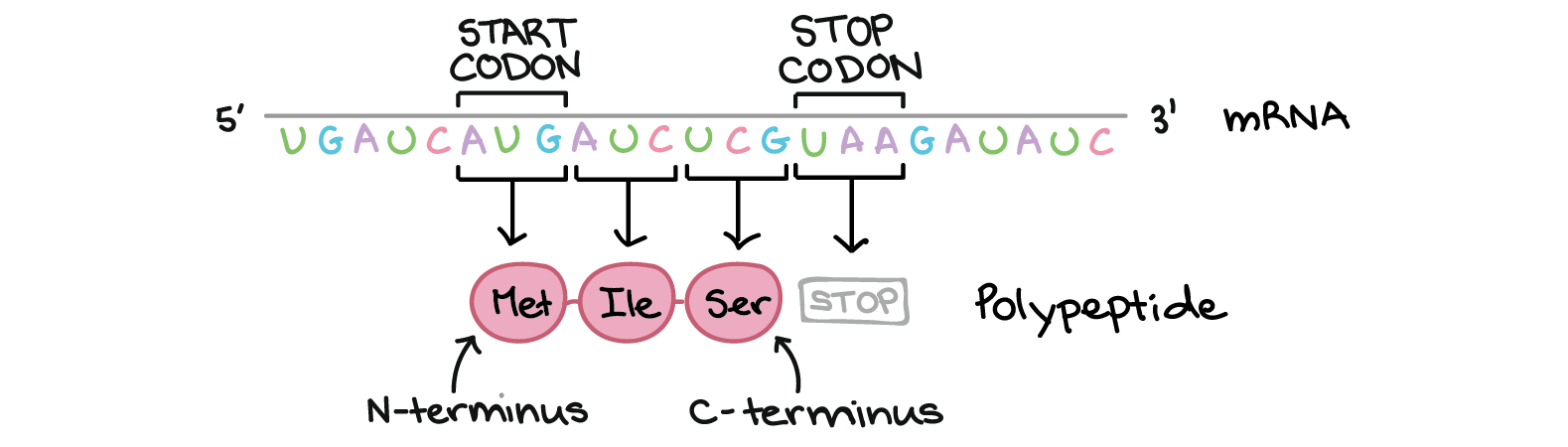
Open reading frame
An open reading frame (ORF) is a continuous stretch of codons that may begin with a start codon and ends at a stop codon and it is the part of a reading frame that has the ability to be translated. When we look an mRNA, there are three ways, how it could be translated: starting in first position, second position or third position. Starting in fourth position is the same as starting
Molecular process of translation
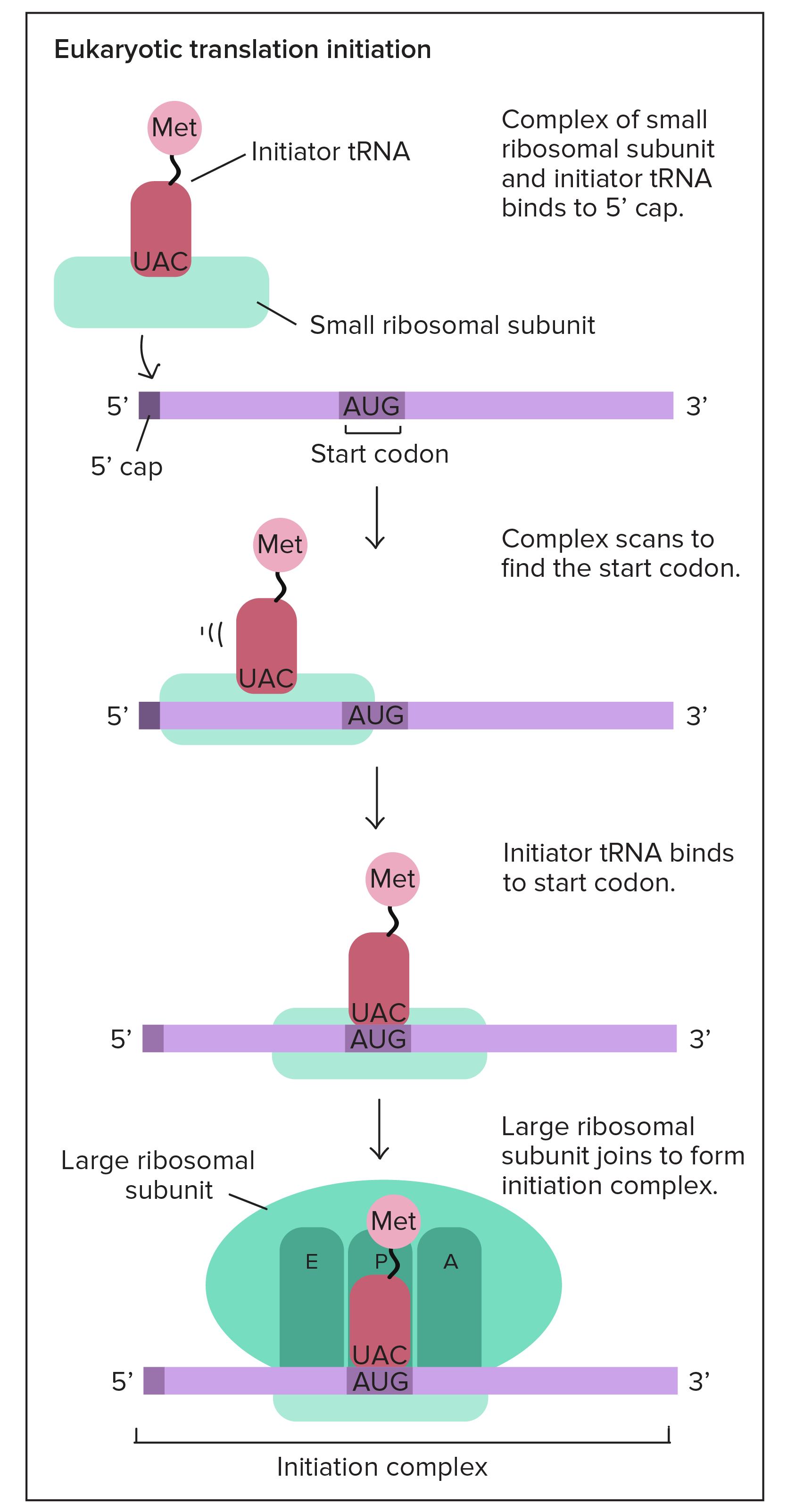
Codons of mRNA are translated into amino acid of protein by tRNA molecules. tRNA molecule contains an anticodon, what is three-letter sequence complementary to the target codon sequence. This tRNA molecule also contains on itself an amino acid corresponding to matched codon. Process of translation is guided by organelle called ribosome. Ribosome is made out of RNA and protein and this organelle has two pieces - small and large subunit. These subunits can exists independently, but they have to come together to do the translation.

Phases of translation
Whole process of translation consists of three phases:
1. Initiation
In the phase of initiation of translation, small subunit of ribosome binds close to the start codon. The next step is that tRNA identifies AUG start codon, binds there and stabilizes binding of small ribosomal subunit to mRNA. After this, large ribosomal subunit can also binds there and make the whole ribosome active.
2. Elongation
During the elongation phase, the ribosome will start moving through mRNA sequence by three nucleotides. Inside the large subunit of ribosome, there are three positions called A (aminoacyl), P (peptidyl) and E (exit). The A is an acceptor side and this is where the new tRNA that identifies the triplet comes in. The P is peptidyl site and the protein (peptide) that is being created is attached here. The E is an exit site and it contains tRNA that already given its amino acid and is ready to be discarded and recycled. The whole job of the ribosome is to create a chemical bond between amino acid in the A position and amino acid in already created chain, and then shift by three nucleotides. This shifting will discard tRNA in E position and create space for new tRNA in A position.

3. Termination
Once the ribosome reaches the end, stop codon is identified by special tRNA. This tRNA is pretty similar to all the other tRNAs, only it doesn’t have an amino acid associated with it. There is no other amino acid to add to the chain and when ribosome moves protein chain is released because there is nothing anchoring it. Two subunits of ribosome get detached and translation ends here.

The whole molecular process of translation is visualised in following video.
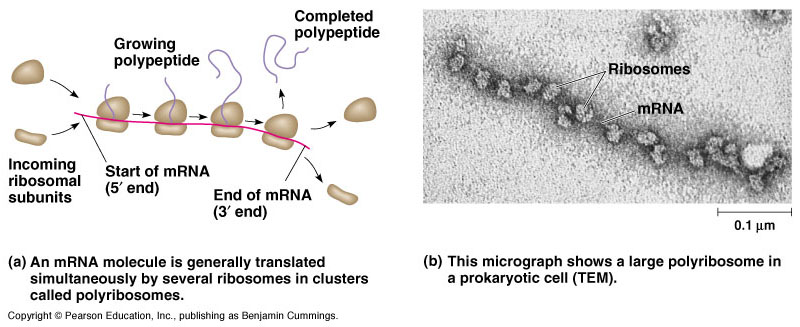
Polyribosome
So far we were talking about one ribosome on one mRNA, but the reality is that mRNA generally gets translated by many ribosomes at the same time. A polyribosome (or polysome or ergosome) is a group of ribosomes bound to an mRNA molecule like “beads” on a “thread”. It consists of a complex of an mRNA molecule and two or more ribosomes that act to translate mRNA instructions into polypeptides.
Quality control
There are two main mechanisms of quality control associated with translation that can message the cell about problem with mRNA that is being actively translated. These two mechanisms will stop translation and send this mRNA to be destroyed and recycled. They have similar ideas, but they are kind of the opposite of each other.
Nonsense-mediated decay
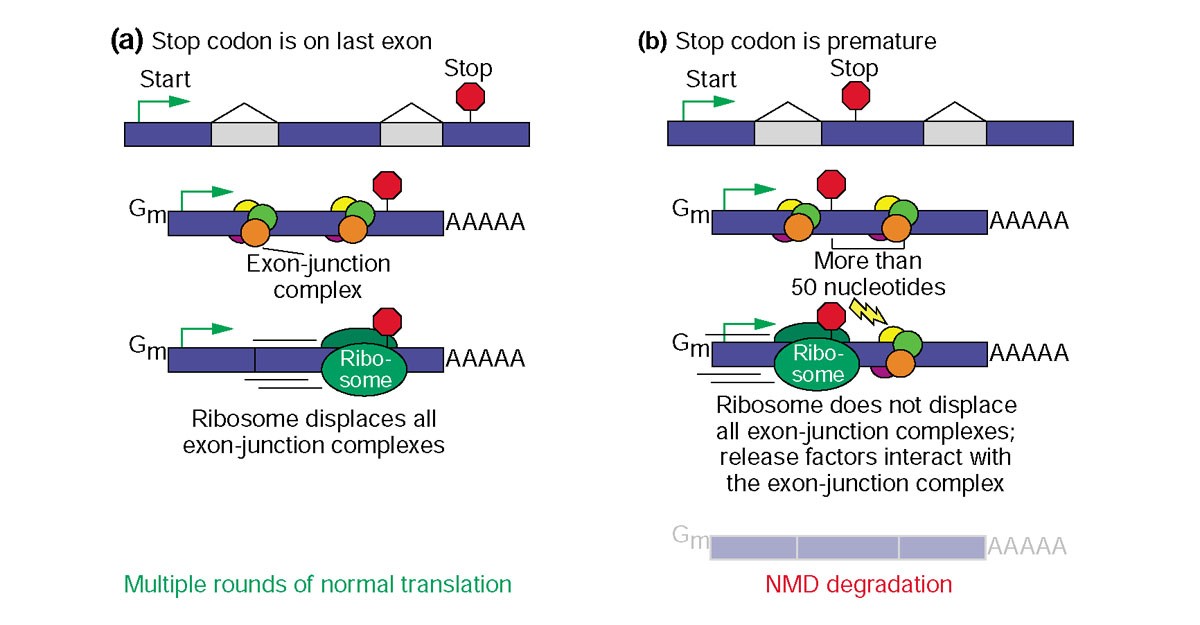
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Premature stop codon causes the protein to be shortened, which may or may not be functional, depending on the severity of what is not translated.
How can translation mechanism know that it reached premature stop codon that shouldn’t be there? After the stop codon there is 3-prime UTR which could be as long as the as the coding sequence, so looking at length of remaining sequence is not helpful.
What the ribosome does, other than creating a protein, is that it clears out all the RNA binding proteins that are bound on the mRNA. One of RNA binding proteins that are bound to mRNA is exon junction complex. When introns are removed from RNA, in process called splicing, this marker protein - exon junction complex - is placed there. Mature mRNA ready to be translated contains exon junction complex on each site where intron was removed. As the ribosome translates the mRNA, it strips the mRNA of these proteins. If some of these proteins have stayed on mRNA after the first round of translation then a nonsense-mediated decay is triggered because it means that the ribosome didn’t reach all the way to the point where the last splicing happened.
Nonstop-mediated decay
When we have a mRNA with mutation in stop codon so it’s not there anymore, ribosome is not released on position of stop codon but continues to translate the rest of mRNA, including 3-prime UTR and poly A tail, and then ribosome get stuck on the end of mRNA. This can be detected by a cell and mRNA is degraded.
Mutations
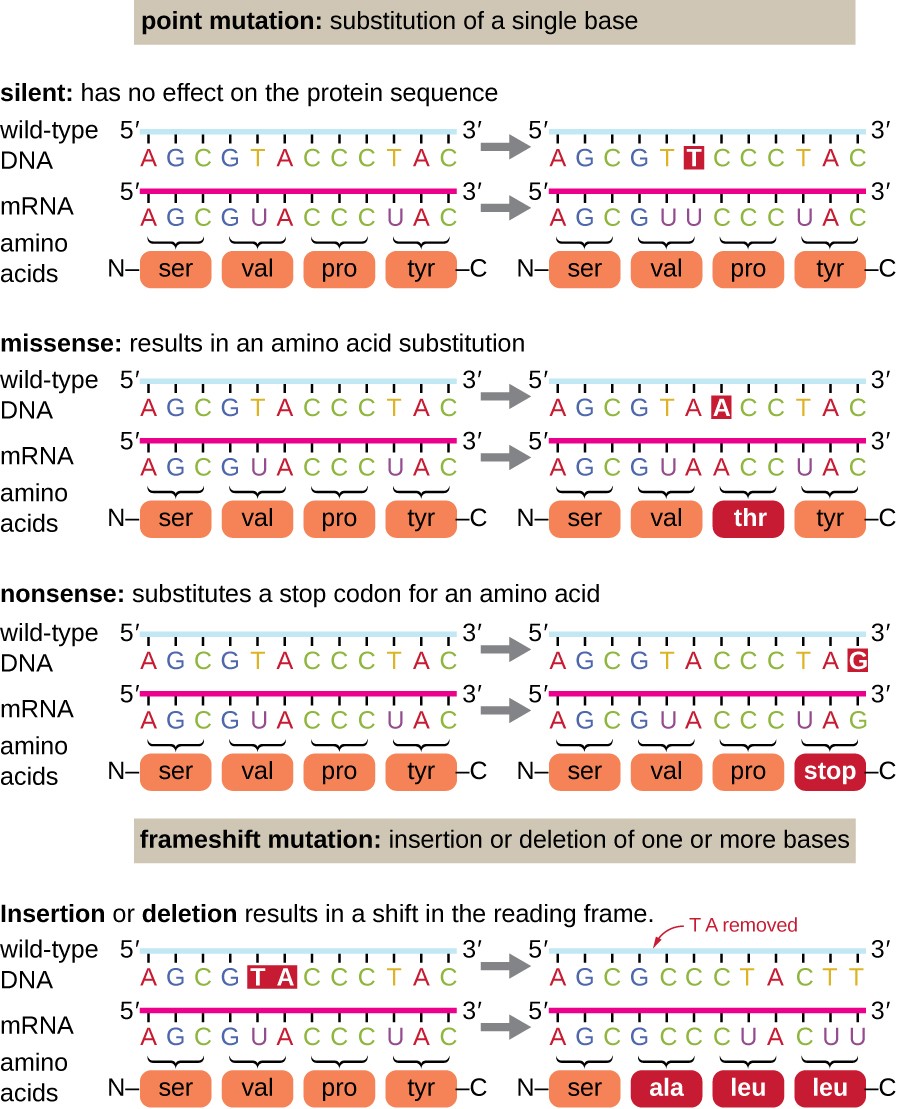
Making copies of DNA and transcribing it into RNA is not a perfect process. There can be changes to genetic informations along the way and we call this changes mutations. There are tree main types of mutations: substitution, insertion and deletion.

These changes are also propagated trough process of translation and they can affect resulting protein. Since multiple codons are encoding the same amino acid, we might get the same amino acid even after mutation, which is called silent mutation. We might get different amino acid or even stop codon instead of amino acid. Insertion and deletion are shifting reading frame what is resulting in change of multiple amino acids.
Single nucleotide polymorphism
Let’s say we have an mRNA coding sequence that is going to make a protein. And let’s say that there’s a mutation that affects a single nucleotide, like a nucleotide changes by instead of being A, it’s going to be a U now. We call this mutation a single nucleotide polymorphism (SNP). This mutation can possibly affect one amino acid - new codon is encoding for different amino acid. The worst outcome could be that it produces a stop codon. Then protein that’s going to be produced will be shorter and it will trigger nonsense-mediated decay.

Frame shift
Bigger changes can happen by an insertion or a deletion mutation - a single nucleotide or a few nucleotides are inserted or deleted. Insertion or deletion of 1 nucleotide shifts everything that is downstream from the shift.
