Transcription
Process of transcription happens in every cell and every organism. Transcription in one of processes of transferring genetic information in cell described in central dogma of molecular biology. It is a process in which DNA is transcribed to RNA.
Personal note
These notes are to some extent similar to notes about genes since transcription is process of creating RNA from gene part of DNA. I will keep duplicate parts in both notes (and just use some references) so they can bring slightly different view and maybe better overall understanding.
DNA exists in double stranded mode (mostly), where both strands are created from four building blocks (four nucleotides - A, C, G and T). These two strands are reversely complementary to each other. Complementary means that every time there is an A in one position there will be a T in the other strand and every time there is a G there will be a C and vice versa. Reversely means that we need to take into account that these strands have an orientation. Strands of DNA are antiparallel - they run parallel to each other but with opposite directionality.
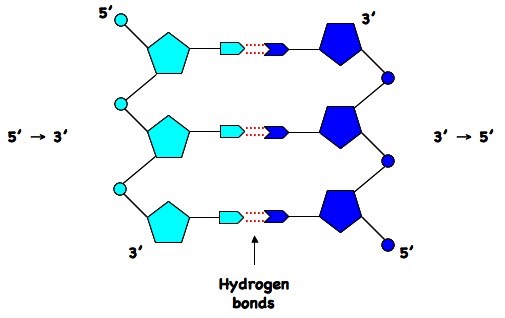
Personal note
This is important to have in mind when working with sequences of DNA (or RNA) in a computer, where we usually have direction of all sequences going from left to right. Let’s say, we have a part of DNA and we are trying to find location on other DNA, where it could bind. In this case we should should reverse our small DNA sequence and then look for a complementarity. Or maybe even better, create a reverse complement of our small sequence and then look for a exact match.
Process of transcription
Since DNA is in double stranded formation and bases are hidden in the middle, DNA needs to open a little before transcription can start so that machinery that is going to do the transcription can get to the right position. Upstream of a gene, there is a regulatory region called promoter. This region can be really close to the beginning of a gene or it can be even few 100 nucleotides upstream of a gene. Certain proteins called transcription factors will come and bind to the promoter. There are special transcription factors that will promote initiation of transcription. They do it by attracting the main protein that is doing the job of transcription that is called polymerase. The most common polymerase in eukaryotic cells is polymerase 2 (POL2) that synthesizes precursors of mRNAs and most sRNA and microRNAs. There are four other known polymerases in eukaryotic cells.
When polymerase gets recruited by transcription factors and get stabilized, it opens a little pocked in double stranded DNA. After this step, it identifies the beginning of a gene - transcription start site - and start moving in a direction of a gene, taking nucleotides from an environment and producing RNA. Visualisation of this whole process is in following video.
RNA is produced in a complementary way. When we say that the gene is somewhere on the DNA, what is actually being read is the opposite strand. The opposite strand is used as a template for the polymerase to create complementary RNA sequence so what comes out is in the same orientation and sequence as the gene.
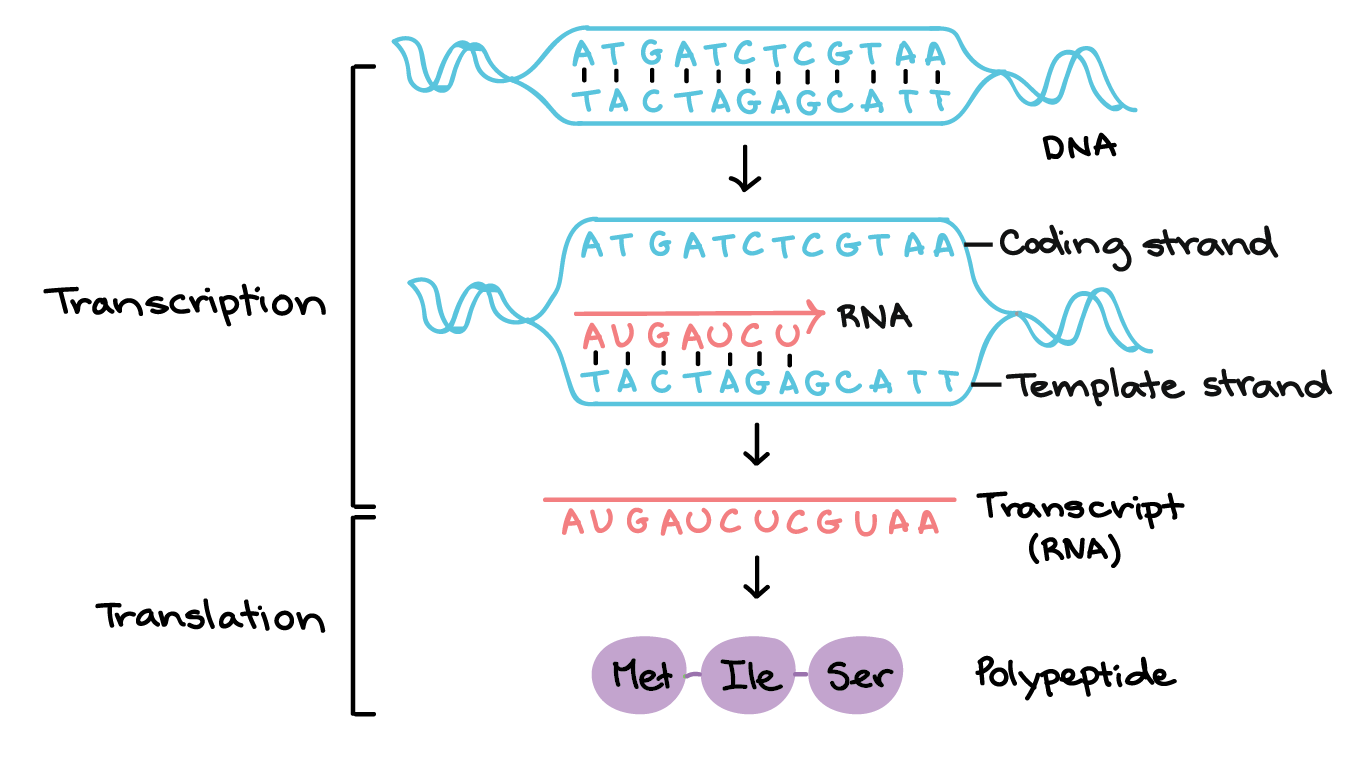
The only difference between the RNA and the DNA gene is that the RNA has the U nucleotide instead of T nucleotides. These two nucleotides are chemically relatively similar.
Post transcriptional RNA modifications
There are some other processes that happen at the same time as transcription, or right after transcription. RNA produced just by transcription is not mature and it is not ready to do its function. There are several modifications that happens to RNA, either co-transcriptionally or right after transcription.
Splicing
RNA produced by transcription is called pre-mRNA and it consists of 5-prime untranslated region (5’ UTR), coding sequence (CDS) and 3-prime untranslated region (3’ UTR). Coding sequence is part that will produce a protein, but not the whole sequence will be used. There are parts called introns which are sequences that are not going to be translated, and they’re actually going to be cut out of the RNA.

Splicing is a process by which introns are removed. This processes is happening thanks to protein-RNA complex called spliceosome. Parts of spliceosome are able to identify specific small sequence motifs in RNA. Spliceosome folds the RNA so that two exons come close together and intron creates loop that can be cut off. It is a very complicated process and there is a lots of regulation. It is very important to get right the edges of the exons (removing one more or one less nucleotide would completely change meaning of resulting RNA). Visualisation of this whole process is in following video.
Removed introns may be degraded, but some of them become functional non-coding RNAs that do other things in a cell. We know for example, that some introns become micro RNAs, which is really interesting way of reusing the cut off parts.
5’ capping
5’ prime capping is relatively simple process. What happens is that first nucleotide of RNA gets chemically modified. The beginning of RNA becomes chemically modified so that it’s identified as stable RNA (it is not just fragment of RNA, intron or something like that). This ensures stability of RNA. There are some proteins whose job is to break up RNA to recycle it. This 5’ cap gives a signal that this RNA is functional and shouldn’t be degraded.
Polyadenylation
Polyadenylation, or Poly(A) tailing, is process that works on 3’ end of RNA. There is AAUAAA sequence on RNA that is identified by some proteins. Part of RNA is cut off and poly-A tail is added to the end of RNA. Poly(A) tail means elongating RNA with many A nucleotides. This poly(A) tail has various functions. It can protect RNA agains degradation by exonuclease. Exonuclease might recognise poly(A) tail and then don’t break down RNA. Or it might start chopping off As from tail, but rest of RNA is still functional and can do its job, before exonuclease gets through tail. There are also some other mechanisms that can identify RNAs that don’t have a poly(A) tail and target them for degradation.
Poly(A) tail is most common for mRNA (messenger RNA - RNA that will produce a protein). But there are also many noncoding RNAs or non protein coding RNAs that are important for a cell but they might not have a poly(a) tail. This has been an issue with many experimental techniques in which a first step of isolating whole RNAs is to use a poly(T) probe that will capture a poly(A) tail. This way we can grab everything that has a poly(A) tail and sequence it. But this approach has a problem that it targets mostly mRNA and not so much non coding RNAs.
Nucleotide modifications
Nucleotide modifications is a process in which certain nucleotides become modified. For example, there can be a cytosine (C) in some location in RNA and it can be modified by adding a methyl group. This modification will slightly change its chemical properties.

RNA-binding proteins
RNA almost never exists by itself. There are many RNA-binding proteins that are in different ways affecting what the RNA is doing. They might be affecting the way it’s folding in space, the way it’s being transported into different places in the cell, they might be shielding it from degradation, they might be degrading it, they might be doing splicing, capping, poly(A) tailing, modifications, they might do quality control of RNA and other things. Slightly scary looking overview of functions of RNA binding proteins is in following image.

Non coding RNA
So far we talked mostly about mRNA which is type of RNA that will eventually produce a protein. Some RNAs will never be translated into protein and they function in a form of RNA. This type of RNAs is called non coding RNAs.
tRNA
tRNA is connected with translation - it is an interface between RNA and protein. On one side, it has a part that identifies a specific sequence of RNA and on the other side, it has corresponding amino acid. These are many different tRNAs for identifying different parts of RNA and carrying different amino acids.
tsRNA
tsRNA comes from tRNA, it is a fragment of tRNA, but it does completely different job that has to do with regulation of RNA life and RNA stability.
rRNA
rRNA or ribosomal RNA is part of a ribosome which is a protein-RNA complex that does translation.
Personal note
Thanks to its properties, rRNA is widely used for bacterial classification - identifying bacterial species. You can read more about it in my Master’s thesis: Bacteria Classification into Taxonomic Categories Based on Properties of 16s rRNA (in Slovak language).
miRNA and siRNA
miRNA (micro RNA) and siRNA (small interfering RNA) are two types of RNA that are very short (20-30 nucleotides). Their job is to identify other RNAs and destroy them. They have a way of identifying their target, binding to it and then attracting protein complex that will destroy this target.
They are useful for defence against viruses and bacteria and other stuff that could come into the cell. They are also useful against cancer. Cell knows what RNAs should be present and which RNAs it needs. So it could have a number of micro RNA’s that are targeting other RNAs that should not be present at that moment. If these RNAs get start getting produced, for whatever reason, they can just identify and destroy them so the cell can keep doing its job even if there’s some small problems with regulation of transcription - what’s being transcribed.
Another important role of these miRNA and siRNA is regulating shifting of programming of a cell. In early development, it is common that after split of a cell, each of daughter cells will do completely different job. These two cells need to very radically and very quickly reprogram themselves. Producing these RNAs that will clear out previous programming so that the new one can start working really fast is useful approach.
lncRNA
This is a big category of various non coding RNAs. It is a general category for all non coding RNAs that are not short - they are above 200 nucleotides in length. So this category is not based on function, but it is based on length. These long non coding RNAs can be catalysts for chemical reactions, they can be structural, they can act as sponges and attract various micro RNAs and then get degraded as a whole.
snRNA
snRNA, or small nuclear RNA, are located in nucleus. Their primary function is in the processing of pre mRNA in the nucleus. They have also been shown to aid in the regulation of transcription factors or RNA polymerase 2, and maintaining the telomeres.
piRNA
Part of job of piRNAs is to control jumping repeat elements. These elements are DNA elements that have evolved into being independent thing of themselves that can copy and paste themselves in different parts of the genome. piRNAs are defence mechanism of a genome against these jumping elements.
Another role of piRNA is localization of RNA. piRNAs can identify sequences and they are used as a kind of glue trap so they can hold necessary RNAs in place while other RNAs are washed away by other cell mechanisms. It’s a good example of nature reusing things that are already there for another purpose.