Genes
In biology, a gene is usually defined as basic unit of heredity and a sequence of nucleotides in DNA or RNA that encodes the synthesis of a gene product - either RNA or protein.
Evolution of gene definition
Genes are central concept in genomics, but it is really hard to define them. We can follow how definition of gene was evolving together with molecular discoveries.
Classical period (mid 18s - early 19s)
During this time, scientists didn’t know about DNA and related stuff. Gene was just some abstract idea. They were something that lived inside the cell and it was able to transport hereditary information from one generation to the next.
NeoClassical period (1940s - 1980s)
This period is after discovering structure of DNA. Gene is defined as part of DNA sequence that acts autonomously and produces protein (trough RNA intermediate). All this is also connected with concept of central dogma of molecular biology, where DNA is transcribed into RNA and then RNA is translated into protein.
Modern period (after 1986)
Modern period is connected with whole genome sequencing. We got much deeper into annotation and understanding of genome. We know that there is a huge amount of genes that don’t make a protein (their final product is RNA). Another discovery was about alternative splicing, which means that the same part of DNA can produce multiple similar (but not the same) proteins.
Today
Today we are in situation where “the gene” cannot be simply defined. But for our purposes we can use this working definition: gene is a genomic locus that produces related transcripts.
Notes on why is hard to define gene
- in addition to protein coding genes, there are many RNA-encoding genes that produce diverse RNA molecules that are not translated to proteins,
- with alternative splicing, different parts of RNA can be translated to create different proteins (from the same part of DNA),
- genes can be separated in pieces, that can be found as separate segments around genome,
- exons of different genes can be members of more then one transcript,
- there are few if any boundaries to transcription,
- there are overlapping genes in genome.
Typical gene
Typical gene is location on DNA (on some chromosome). Start point of gene is called transcription start site (TSS) and it is the first part where RNA start getting produced. First part of a gene is called 5-prime UTR, where UTR stands for untranslated region. This means that first part of RNA that get produced will not be translated to protein. This part is kind of like control sequence. There is a point where the untranslated region stops and coding sequence (CDS) starts. Coding means protein coding - this is the part that will produce protein. After CDS, there is 3-prime UTR that also will not be translated to protein. This direction is always the same - 5-prime is at the beginning and 3-prime is at the end.
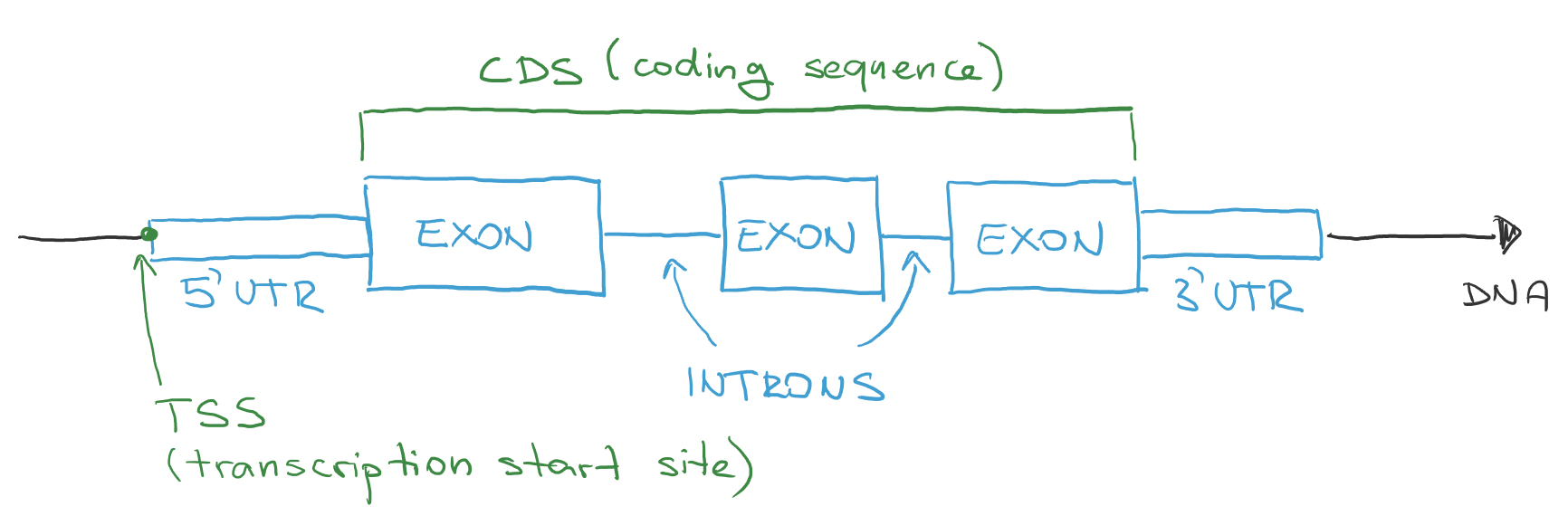
Coding sequence consists of two types of sequences: exons and introns. Exons are parts that will make the protein in the end. Introns are untranslated sequences that are between exons and they will not make a protein. They are removed from the RNA in a process called splicing.
Non-coding gene
This typical gene is an example of protein coding gene - goal of that gene is to eventually get translated into protein. But there are also many non-coding genes and function of these genes essentially happen at the level of RNA. For RNA to function properly, it is necessary that it fold into specific shape. Shape of RNA is important for its function. RNA is just single chain of nucleotides (DNA has two chains that are bound together) but these nucleotides are still capable of creating pairings. RNA chain can fold and create local pairings with itself.

Gene splicing
Right after transcription, RNA consists of 5-prime UTR, exons, introns and 3-prime UTR. And it also might have a long string of A’s at the end. They are called poly-A tail and they are added to the RNA after transcription from DNA. This is a code for a cell that this molecule is RNA that needs to be translated into protein. It is added to the end of RNA only after successful transcription. Damaged transcripts don’t have this poly-A code and they can be destroyed by cell.
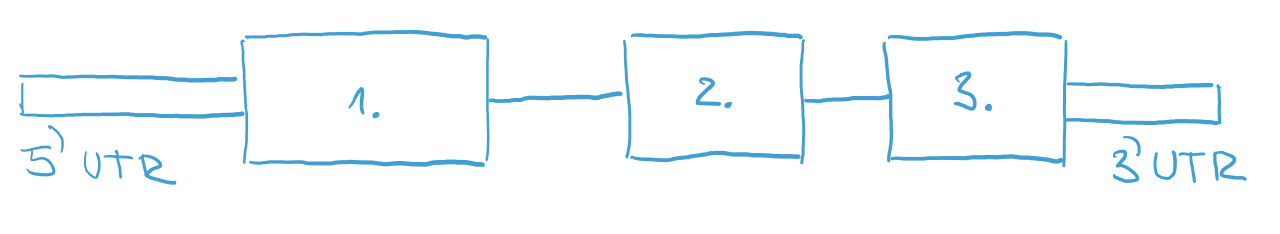
Another modification of RNA is splicing. During splicing, introns are removed from RNA.
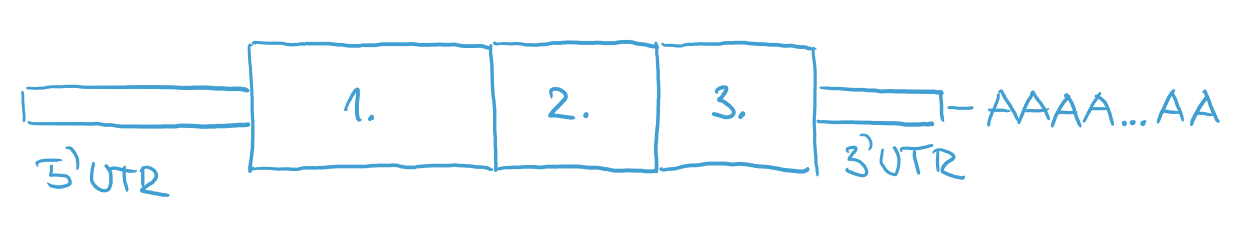
After this step, RNA is ready to go into translation to produce a protein.
However, RNA is almost always instantly bound by various proteins and are all almost always associated with proteins that are helping the RNA to do its job. These various proteins are called RNA-binding proteins and they do many different jobs. For example, there are RNA-binding proteins that control how splicing will happen. There are other RNA-binding proteins that control localisation of RNA in cell - this specific RNA-binding protein will have a way use some kind of transport mechanism to bring this RNA to the position where it needs to be. Other RNA-binding proteins control life cycle of RNA. They control how long will this RNA survive and if it will get degraded.
Alternative splicing
A very common thing that happens in almost every gene is called alternative splicing.
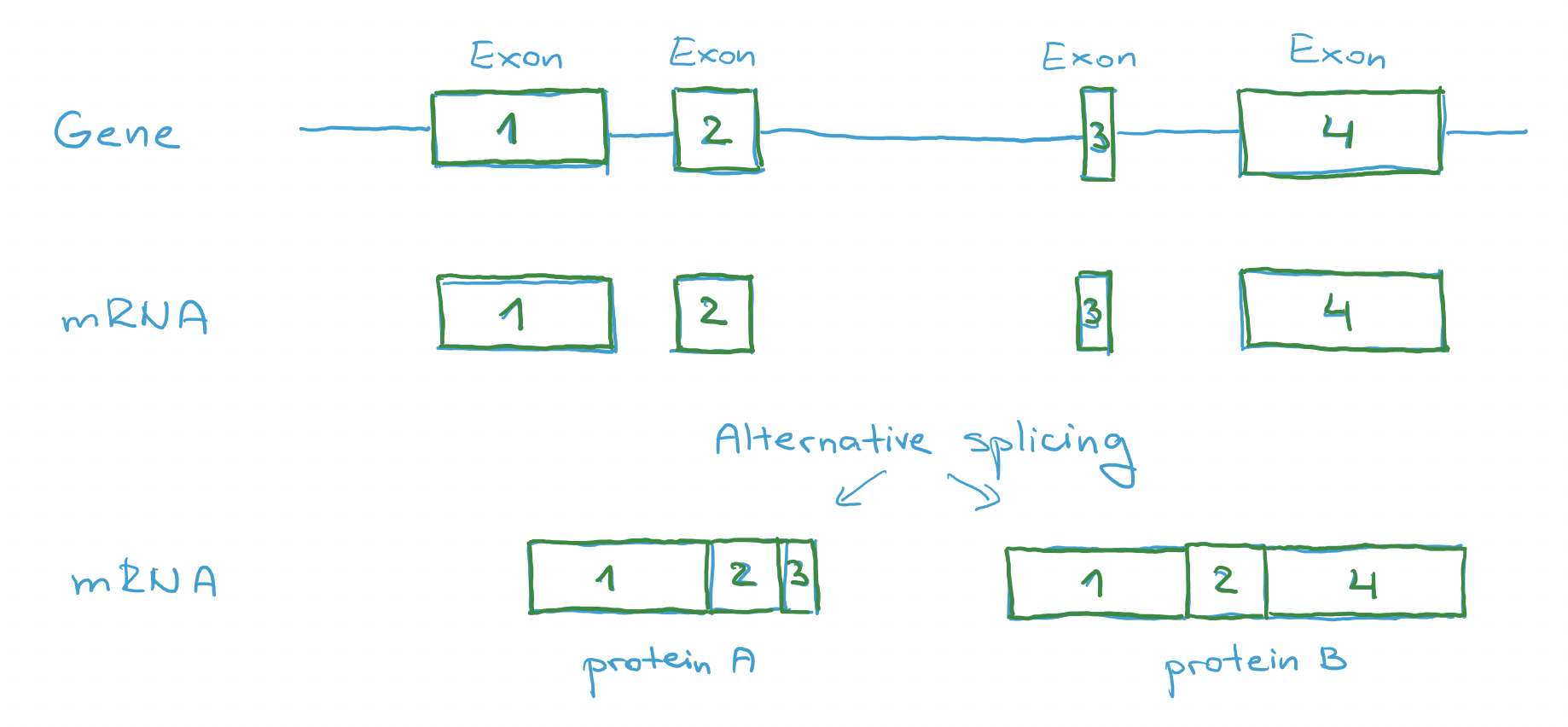
Removal of introns is not exact. Some introns might stay in RNA and some exons might be removed. This is used to make group of proteins that might do slightly different job, but we still call this proteins with the same name and we still call the gene with the same name.
Regulatory regions
Transcription of a gene is regulated by DNA regions that are located outside of a gene.

Little bit before the gene, there is an area in the DNA that is called the promoter region. The promoter region is kind of like a control panel that controls transcription of a gene. This promoter region contains specific sequences that can be recognised by some proteins that exists in cells that are called transcription factors. Various different transcription factors can bind to this promoter region. Based on the combination of transcription factors that bind the promoter region, they can activate or suppress transcription of a gene. A big question in biology is when given a transcription factor and and a sequence of DNA, what part of the sequence does the transcription factor recognise and will this transcription factor bind to this sequence? This is an important question because the better we model these interactions, the more we can understand the regulation of genes.
Another regulatory region of a gene is enhancer. Enhancer is similar to promoter (it also controls expression level of a gene), but it can be really far away from the gene that it controls. There could be one million bases between enhancer and gene, so this is a long distance interaction. Gene can be associated with several of these enhancer.